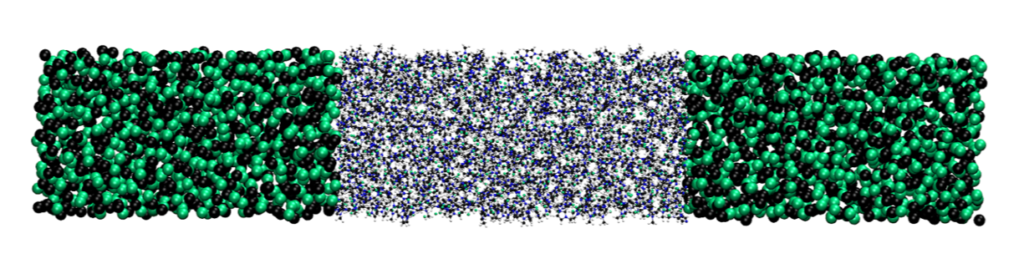
The Grand Canonical Adaptive resolution scheme (GC-AdResS) gives a methodological description to partition a simulation box into different regions with different degrees of accuracy. For more details on the theory see Refs. [1,2,3].
In the context of an E-CAM pilot project focused on the development of the GC-AdResS scheme, an updated version of GC-AdResS was built and implemented in GROMACS, as reported in https://aip.scitation.org/doi/10.1063/1.5031206 (open access version: https://arxiv.org/abs/1806.09870). The main goal of the project is to develop a library or recipe with which GC-AdResS can be implemented in any Classical MD Code.
The current implementation of GC- AdResS in GROMACS has several performance problems. We know that the main performance loss of AdResS simulations in GROMACS is in the neighbouring list search and the generic serial force calculation linking the atomistic (AT) and coarse grained (CG) forces together via a smooth weighting function. Thus, to remove the bottleneck with respect to performance and a hindrance regarding the easy/general implementation into other codes and eliminate the non optimized force calculation, we had to change the neighbourlist search. This lead to a considerable speed up of the code. Furthermore it decouples the method directly from the core of any MD code, which does not hinder the performance and makes the scheme hardware independent[4].
This module presents a very straight forward way to implement a new partitioning scheme in GROMACS . And this solves two problems which affect the performance, the neighborlist search and the generic force kernel.
Information about module purpose, background information, software installation, testing and a link to the source code, can be found in our E-CAM software Library here.
E-CAM Deliverables D4.3[5] and D4.4[6] present more modules developed in the context of this pilot project.
References
[1] L. Delle Site and M. Praprotnik, “Molecular Systems with Open Boundaries: Theory and Simulation,” Phys. Rep., vol. 693, pp. 1–56, 2017
[2] H.Wang, C. Schütte, and L.Delle Site, “Adaptive Resolution Simulation (AdResS): A Smooth Thermodynamic and Structural Transition fromAtomistic to Coarse Grained Resolution and Vice Versa in a Grand Canonical Fashion,” J. Chem. Theory Comput., vol. 8, pp. 2878–2887, 2012
[3] H. Wang, C. Hartmann, C. Schütte, and L. Delle Site, “Grand-Canonical-Like Molecular-Dynamics Simulations by Using an Adaptive-Resolution Technique,” Phys. Rev. X, vol. 3, p. 011018, 2013
[4] C. Krekeler, A. Agarwal, C. Junghans, M. Prapotnik and L. Delle Site, “Adaptive resolution molecular dynamics technique: Down to the essential”, J. Chem. Phys. 149, 024104
[5] B. Duenweg, J. Castagna, S. Chiacchera, H. Kobayashi, and C. Krekeler, “D4.3: Meso– and multi–scale modelling E-CAM modules II”, March 2018 . [Online]. Available: https://doi.org/10.5281/zenodo.1210075
[6] B. Duenweg, J. Castagna, S. Chiacchera, and C. Krekeler, “D4.4: Meso– and multi–scale modelling E-CAM modules III”, Jan 2019 . [Online]. Available: https://doi.org/10.5281/zenodo.2555012