Abstract
In this conversation with Andreas Singraber, post-doc in E-CAM until last month, we will discover the ensemble of his work to expand the Neural Network Potential (NNP) Package n2p2 and to improve user accessibility to the code via the LAMMPS package. Andreas will talk about new tools that he developed during his E-CAM pilot project, that can provide valuable input for future developments of NNP based coarse-grained models. He will describe how E-CAM has impacted his career and led him to recently integrate a software company as a scientific software engineer.
With Dr. Andreas Singraber, Vienna Ab initio Simulation Package (VASP)
Could you please give us an overview of the practical problems that initially motivated your pilot project in E-CAM?
The pilot project topic “Implementation of neural network potentials for coarse-grained models” was inspired by recent publications showing that machine learning potentials can in principle be used as a basis for creating coarse-grained (CG) models. Although coarse-graining is a well-known technique to overcome limitations of accessible time scales and system sizes of a fully-atomistic description, its combination with machine learning was mostly uncharted territory. In particular, no one had tried to use high-dimensional neural network potentials in this context.
What was the initial goal ?
The idea behind the project was to create neural network potential (NNP) based CG models of more complex systems than the ones that were presented in the proof of concept publications. More specifically, the focus was on dendrimer-like DNA molecules which were definitely an ambitious target. Although multiple coarse-graining pathways looked feasible it was unclear whether the approach would work for such large molecules. At the same time the necessary software tools would be written and documented in order to lower the barrier for future users to apply the method. In addition, as it was expected that training and application of NNP based CG models would further increase the computational demand, strategies to increase the performance of existing software would be sought and implemented. As a side effect these general improvements would ideally also be beneficial for conventional NNPs.
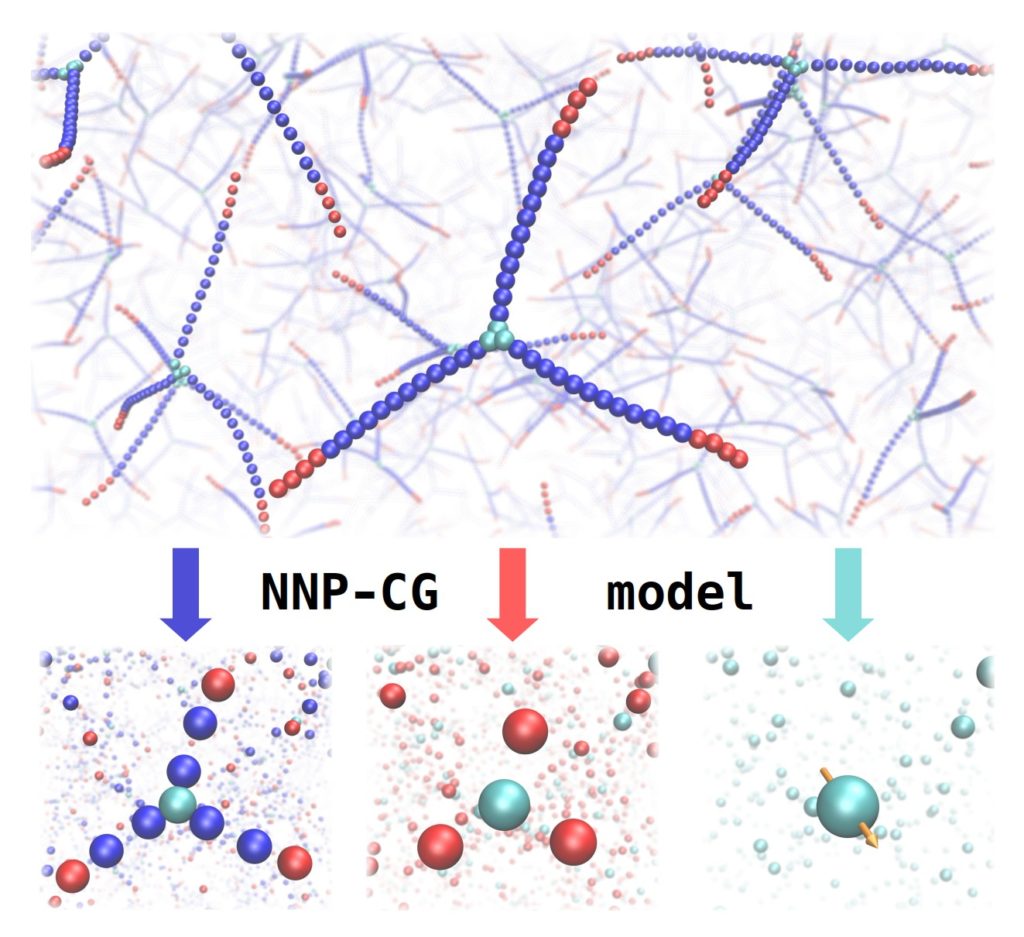
Most Coarse Grained methods use some form of bead representation of a system, where many atoms are mapped into each bead. Is your approach similar to that? And if this is the case, how are the NNP atomic environment descriptors related to the beads?
Yes, the common approach of a bead representation can be the basis for creating a NNP based CG model. For example, a simple toy model I tested was to represent a water molecule by a bead located at the centre of mass. As was already shown in the literature for other machine learning potentials this works reasonably well. In such a simple case the usual descriptors (symmetry functions) can be used without modification, even multiple bead types would work by assigning them to different elements. I tried a “multi-bead-type” approach for dendrimer-like DNA molecules but without much success. For more complex molecules a different representation may be better suited. For example, a bead at the center of mass could be equipped with an additional vector determining the orientation of a disk-shaped molecule. However, for such a representation there are no descriptors available yet.
How does the neural network training work scale with the number of beads or basis functions?
On the one hand, if the coarse-grained representation reduces the number of different particle types, then this also relaxes requirements on the NNP, in particular the number of descriptors decreases. However, there are important, complicating differences with respect to conventional training when it comes to NNP based CG models. First, the NNP is trained to reproduce the free energy surface (instead of the potential energy surface) and therefore only instantaneous collective forces are available as training patterns. Moreover, these CG forces must be sampled from equilibrium MD simulations at the fully-atomistic level which may result in huge training data sets. Also, it may be necessary to incorporate an additional conventional baseline potential to avoid sampling problems related to unphysical structures with close-by CG beads.
Could you outline the main results from your pilot project to date?
Starting with the scientific perspective probably the n2p2 – CG descriptor analysis tools gave the most important insight into the expected accuracy of NNP based CG models. The idea behind this analysis was to get a rough picture of how well the combination of a coarse-grained representation and environment descriptors used in machine learning will work before even starting to train a model. In practice this is done by looking at correlations between forces in the training data and the corresponding descriptors with the help of a clustering algorithm and statistical tests. The descriptor analysis is also applicable to conventional NNPs and is documented in great detail for future users.
On the performance side there were significant improvements regarding NNP training. Training of CG models requires large training data sets where the amounts of memory required could easily reach several hundreds of gigabytes in the previously existing implementation. With an improved memory management it was possible to reduce this by approximately 30 to 50%, depending of course on the studied system. A little less related, but still potentially useful for CG model generation, is the development and implementation of a new class of atomic environment descriptors in n2p2 called polynomial symmetry functions [1]. These are avoiding the computational overhead from the functional form of conventional Behler-Parrinello type symmetry functions and provide a significant speedup without loss of model accuracy.
The aforementioned developments are based on and integrated in n2p2, a software package for training and application of high-dimensional neural network potentials: https://compphysvienna.github.io/n2p2/index.html. To get an overview of the individual software developments it is best to visit the E-CAM’s software library section dedicated to n2p2 modules at https://e-cam.readthedocs.io/en/latest/Classical-MD-Modules/index.html#n2p2
What challenges did you face?
While I could confirm that the general idea of creating NNP based CG models can be successful I must admit that the ultimate goal of a CG model for dendrimer-like DNA molecules was too ambitious. It is not yet clear what caused the training procedures to fail but given the results from the descriptor analysis I suspect that the developed CG representations were not appropriate and hence not enough useful information could be presented during the training stage. Nevertheless, the new tools and other software improvements will still prove valuable in future developments of NNP based CG models.
What other achievements did you realise during your time with E-CAM?
Overall the capabilities and performance of n2p2 improved significantly during my time with E-CAM. There were many little enhancements, each code change alone maybe too small to be mentioned in a separate E-CAM module but after crunching the numbers I found that around 35% of today’s n2p2 codebase was added during my E-CAM project. In the last two years 23 pull requests were merged and over 60 issues reported by users (via mail or Github) were resolved. One of the major advantages of n2p2 from the user’s perspective is the interface to LAMMPS which allows to run massively parallelised molecular dynamics simulations with pre-trained neural network potentials. In the near future this connection to LAMMPS will be further strengthened because I recently started a pull request to integrate n2p2 parts directly in LAMMPS as a user contributed package. Having the n2p2/LAMMPS interface description directly available in the LAMMPS online documentation will increase the visibility and broaden the user base. I would like to point out another major development still in progress: the implementation of fourth-generation neural network potentials (4G-HDNNPs, see also [2]). In this new approach Behler and co-workers introduce a new type of neural network potential capable of dealing with long range interactions. A clever combination of a second neural network and a global charge equilibration scheme allows the application of machine learning potentials in molecular systems with charge transfer. Although this is still work in progress (together with collaborators), a lot of changes modularising n2p2 were made in preparation for this new method during my time in E-CAM.
What is the potential impact of your work in the community and in more broader terms, in the society?
Computer simulations have become an important tool in many fields of science, with applications ranging from drug development to the design of new materials. Nevertheless, the predictive power of simulations depends crucially on the model that is used to describe the atomic interactions. Electronic structure methods like density functional theory (DFT) provide an accurate description of those interactions, however at a high computational cost, which prevents the application of these potentials in molecular dynamics or Monte Carlo simulations to study more complex systems. Machine learning (ML) potentials appear as an alternative approach to describing atomic interactions, as they combine the accuracy of quantum mechanical electronic structure calculations with the efficiency of simple empirical potentials. Among the different types of ML potentials developed to date, Neural Network potentials are amongst the most used, and are the ones used in this work. The societal benefit of this work is therefore increasing the availability of ready-to-use and scalable software for high-dimensional neural network potentials in computational physics and chemistry, and thus speeding up the development of new drugs and help in the design of high-tech materials, among many other applications.
Is your work being used by other groups and in other activities ? Does your work have an impact on HPC?
Yes, the performance improvements mentioned above and my work on n2p2 in general during my time with E-CAM is certainly beneficial for many n2p2 users. It is not easy to estimate how many people are actually using the software at the moment but Github statistics shows a good number of “unique” visitors per day. Also, there are currently three development projects where other groups are involved and I only have an assisting role. One I would like to mention in particular because it is related to HPC. A group at the LLNL (Lawrence Livermore National Laboratory) created an interface between n2p2 and their CabanaMD software which is an experimental MD code using the Kokkos performance portability library. They could already show that with some modifications to n2p2 this combination scales up to two million atoms on the GPU and 21 million atoms on a single CPU node. The next step is to port this proof-of-concept code to LAMMPS allowing users to run MD simulations with NNPs also on the GPU which is a very exciting perspective.
Is there a potential industrial interest?
Yes, there is potential interest of commercial software companies to integrate n2p2 in their workflow.
What follow up/ future developments would you visualise for your work?
I am following closely the development of the Kokkos port mentioned before and I hope that soon there will be a scalable GPU implementation of neural network potentials in LAMMPS. Also, I am confident that we will have a suitable parallel implementation of 4G-HDNNPs available in n2p2/LAMMPS which would be a very helpful tool in computational physics and chemistry when dealing with long range interactions and charge transfer. In general, I hope that E-CAM and similar projects around the world will help scientists to increasingly use modern tools for software development.
Will E-CAM have an impact on your career ?
E-CAM already had a huge impact on my career! I came in contact with E-CAM as a PhD student in computational physics participating in two Extended Software Development Workshops workshops. Mostly untrained in modern software development tools I learned there all the tools of the trade on how to contribute to big codebases and I became curious to dig deeper into state-of-the-art scientific programming. This heavily influenced the focus during my PhD studies and ultimately led to the first release of n2p2. I am very thankful that I could continue in this direction when I joined E-CAM as a postdoc in 2019. I always considered it a very important aspect of my work in E-CAM to share knowledge with Master and PhD students I worked with because in my opinion universities do not yet include enough software development topics in their curricula. I assume that many people in E-CAM have the same experience and in that sense I believe that the impact of E-CAM as a whole is much higher than what can be expressed in reports and on websites. Regarding my personal future I am very happy that I recently joined the VASP software company as a scientific software engineer where I will certainly profit a lot from my training in E-CAM.
References
[1] Bircher, M. P.; Singraber, A.; Dellago, C. “Improved Description of Atomic Environments Using Low-Cost Polynomial Functions with Compact Support”. Submitted. arXiv:2010.14414
[2] Ko, T. W.; Finkler, J. A.; Goedecker, S.; Behler J. “A fourth-generation high-dimensional neural network potential with accurate electrostatics including non-local charge transfer”, Nat Commun 12, 398 (2021). https://doi.org/10.1038/s41467-020-20427-2
