Dr. Jony Castagna, Science and Technology Facilities Council, United Kingdom
Abstract
Jony Castagna recounts his transition from industry scientist to research software developer at the STFC, his E-CAM rewrite of DL_MESO allowing the simulation of billion atom systems on thousands of GPGPUs, and his latest role as Nvidia ambassador focused on machine learning.
Jony, can you tell us how you came to work on the E-CAM project and what you were doing before?
My background is in Computational Fluid Dynamic (CFD), and I worked for many years in London as a computational scientist for an Oil & Gas industry. I joined STFC – Hartree Centre in 2016 and E-CAM was my first project. E-CAM offered an opportunity to work in a new and more academic and fundamental research environment.
What is your role in E-CAM?
My role is as research software developer, which consists mainly in supporting the E-CAM Postdoctoral Researchers in developing their software modules, benchmarking available codes and contribute to the deliverables of the several work-packages. This includes the work described here, in co-designing DL_MESO to run on GPUs.
What is DL_MESO and why was it important to port it to massively parallel computing platforms?
DL_MESO is a software package for mesoscale simulations developed by M. Seaton at the Hartree Centre [1, 2]. It is basically made of two software components: a Lattice Boltzmann method solver, which uses the Lattice Boltzmann equation discretize on a lattice (2D or 3D) to simulate the fluid dynamic effects of complex multiphase systems; and a Dissipative Particle Dynamics (DPD) solver based on particle method where a soft potential, together with a coupled dissipation and stochastic forces, allows the use of Molecular Dynamics but with a larger time step.
The need to port DL_MESO to massively parallel computing platforms arose because often real systems are made of millions of beads (each bead representing a group of molecules) and small clusters are usually not sufficient to obtain results in brief time. Moreover, with the advent of hybrid architectures, updating the code is becoming an important software engineering step to allow scientist to continue their work on such systems.
How well were you able to improve the scaling performance of DL_MESO with multiple GPGPU’s, and as a consequence, how large a system can you now treat?
The current multi-GPU version of DL_MESO scales with an 85% efficiency up to 2048 GPUs equivalent to about 10 petaflops of performance double precision (see Fig. 1 reproduced from E-CAM Deliverable 7.6[3]). This allows the simulation of very large systems like a phase mixture with 1.8 billion particles (Fig. 2). The performance has been obtained using the PRACE resource Piz Daint supercomputer from CSCS.
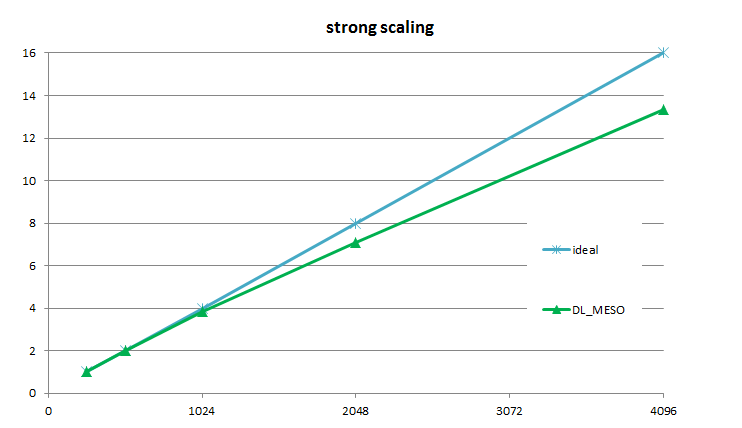
Figure 1. Strong scaling efficiency of DL_MESO versus the number of GPGPU for a simulation of a complex mixed phase system consisting of 1.8 billion atoms.
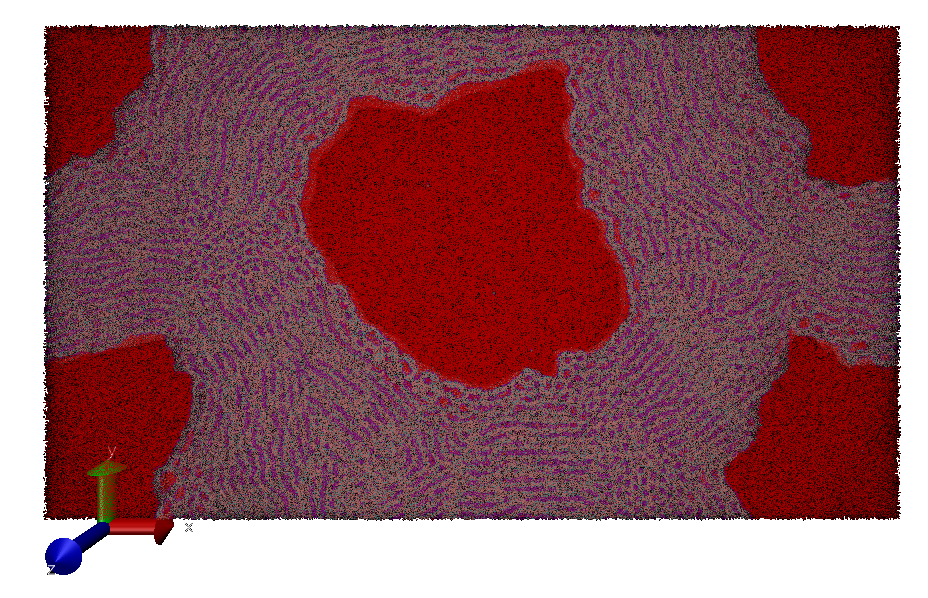
What are the sorts of practical problems that motivated these developments, and what is the interest from industry (in particular IBM and Unilever) ?
DPD has the intrinsic capability to conserve hydrodynamic behavior, which means it reproduces fluid dynamic effects when a large number of beads is used. The use of massively parallel computing allows the simulation of complex phenomena like shear banding in surfactants and ternary systems present in many personal care, nutrition, and hygiene products. DL_MESO has been used intensively by IBM Research UK and Unilever and there is a long history of collaboration with Hartree Centre still going on.
Are there some examples of the power of DL_MESO to simulate continuum problems with difficult boundary conditions, etc., where standard continuum approaches fail?
Yes. One good example is the polymer melt simulation. Realistic polymers typically are notoriously very large macromolecules, and their modeling in industrial manufacturing processes, where fluid dynamic effects like extrusion exist, is a very challenging task. Traditional CFD solvers fail to describe well the complex interface and interactions between polymers. DPD represents the ideal approach for such systems.
What were the particular challenges to porting DL_MESO to GPUs? You started by an implementation on a single GPU and only afterwards ported it to multi-GPUs. Was that needed?
The main challenge has been to adapt the numerical algorithm implemented in the serial version to the multithread GPU architecture. This required mainly a reorganization of the memory layout to guarantee coalescent access and take advantage of the extreme parallelism provided by the accelerator. The single GPU version was developed first, optimized and then extended to multi-GPU capability based on MPI library and a typical domain decomposition approach.
We know you are adding functionalities to the GPU version of DL_MESO, such as electrostatics and bond forces. Why is that important?
Electrostatic forces are very common in real systems, they allow the simulation of complex products where charges are distributed across the beads creating polarization effects like those in a molecule of water. However, these are long-range interactions and special methods like Ewald Summation and Smooth Particle Ewald Mesh are needed to fully compute their effects. They represent a challenge from numerical implementation due to their high computational cost and difficulties they present to parallelization.
Where can the reader find documentation about the software developments that you have been doing in DL_MESO?
Mainly on the E-CAM modules dedicated to DL_MESO that have been reported on Deliverables 4.4[4] and 7.6[3], and also on the E-CAM software repository here .
Did your work with E-CAM, on the porting of DL_MESO to GPUs, opened doors to you in some sense?
Yes. IBM Research UK has shown interest in the multi-GPU version of the code for their studies on multiphase systems and Formeric, a spin-off company of STFC, is planning to use it as the back end of their products for mesoscale simulations.
Recently, you have also been nominated as an NVidia Ambassador. How did that happen?
We have a regular collaboration with NVidia, not only through the Nvidia Deep Learning Institute (DLI) for dissemination and tutorials, but also for optimization in porting software to multi-GPU as well as Deep Learning applications applied mainly to computer vision industrial problems. This is how I got the Nvidia DLI Ambassador status in October 2018. It is being a great experience and an exciting opportunity.
What would you like to do next?
The Nvidia Ambassador experience in Deep Learning opened a new exciting opportunity in the so-called Naive Science: the idea is to use neural networks for replacing traditional computational science solvers. A Neural Network can be trained using real or simulated data and then used to predict new properties of molecules or fluid dynamic behaviour in different systems. This will speed up the simulation by a couple of orders of magnitude as well as avoiding complex modeling based on the use of ad hoc parameters that are often difficult to determine.
References
[1] http://www.cse.clrc.ac.uk/ccg/software/DL_MESO/
[2] M. A. Seaton, R. L. Anderson, S.Metz, and W. Smith, “DL_MESO: highly scalable mesoscale simulations,”Molecular Simulation, vol. 39, no. 10, pp. 796–821, Sep. 2013.
[3] Alan O’Cais, & Jony Castagna. (2019). E-CAM Software Porting and Benchmarking Data III (Version 1.0). Available in Zenodo: https://doi.org/10.5281/zenodo.2656216
[4] Silvia Chiacchiera, Jony Castagna, & Christian Krekeler. (2019). D4.4: Meso- and multi-scale modelling E-CAM modules III (Version 1.0). Available in Zenodo: https://doi.org/10.5281/zenodo.2555012
