Dr. Donal Mackernan, University College Dublin
Abstract
The power of advanced simulation combined with statistical theory , experimental know-how and high performance computing is used to design a protein based molecular switch sensor with remarkable sensitivity and significant industry potential. The sensor technology has applications across commercial markets including diagnostics, immuno-chemistry, and therapeutics.
Introduction
A well-known scientific colleague declared, with a certain pride, that he was happy to work on anything that could be cast as a particle, provided he was paid by a sufficiently interesting question. Given his provocative personality the remark did not surprise me. A few years later I find myself astonished that I am now working on a project with possibly very significant real applications for human health, reading papers in molecular biology and immunology, and in the early stage of preparing the business case for a start-up. And I smile remembering the declaration.
The story started by my helping out an enthusiastic PhD student with a strong background in biology but relatively little experience of mathematical methods. He wanted to design bio-sensors. My background is rooted in statistical mechanics and dynamical systems theory, and simulation, where notions of instability, metastability, and the application of bias to accelerate sampling are standard. I knew little or no biology, and biophysics talks simply bewildered me. And yet we started to work together. Biosensors to work well need to be accurate, which generally means that the signal to noise ratio associated with measurement should ideally be high, i.e. false positive and false negative predictions should be rare. One day, I suggested, using a simple model, that in principle we could use bias to maximise the signal to noise ratio. At the time I was in between contracts, looking for work, and spending a lot of time at a UK based patent law firm, studying patent law. That spell in limbo, although it did not seem so at the time, would turn out to be decisive. After about a year, the situation improved and we were able to return to the project, and sometime later, when we had shown that the basic idea did indeed work, I realized what we had was probably patentable.
We had, in effect, used rational computer-based design to develop a biosensor platform, in effect a very smart molecular switch, that could be combined with biomarker sensing units such as antibody fragments, and flagging units such as fluorescent proteins. One of the most novel ideas within the switch was that bias and instability could be combined to create remarkably sensitive and flexible sensors, where all of the complexity of the sensor was built into a single protein that could, in principle, be built using genetic engineering, and expressed in cells, or used in assays. Moreover, due to its nanoscale, the molecular switch could be used in applications where the ability to sense with high precision target molecules and turn on or off protein interactions accordingly are useful, such as diagnostics, visualization of cellular processes, and therapeutics. The spell in limbo had at least taught me the basics of how to draft a patent, for which the PhD student and I were the declared inventors, and later benefit from the considerable support from our university technology transfer office.
The role of E-CAM
Developing and optimizing the sensor for different bio-marker targets and environmental conditions using rational design and simulation is very demanding, and requires many of the methods that are central to E-CAM work-packages 1 and 4; that is rare-event methods and mesoscale/multiscale modelling respectively. A sensor, to be useful, must function inside the cytoplasm of a living cell, or in bodily fluids such as blood serum within a suitable device. This means that account must be made of the physiological environment, in particular, the ambient conditions of salinity (salt), pH, hydration and viscosity of the solvent. This theme is actually part of a UCD E-CAM pilot project in the more general context of food and pharmaceutical proteins. In addition, actual amino acid sequence comprising the sensor in general need to be mutated during the optimization process, which entail changes in free energy and kinetic properties.
Also, the systems that need to be simulated, although small on the scale of the cell, still consist of between 400 and 1000 amino acids (depending on each case), and when the solvent is included around a million atoms. This means that methodological cunning and significant HPC resources are required. Therefore, there is great scope for the application of E-CAM know-how and software in the future development of these sensors. So what had we come up with?
Technology Description
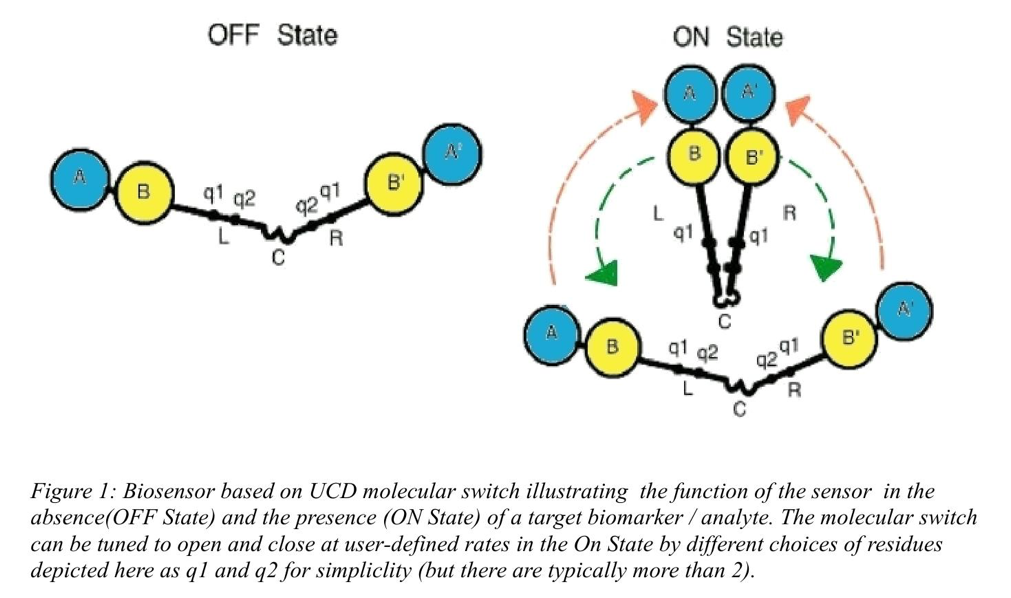
The switch technology is illustrated in Figure 1 and consists of a hinge-like protein fused to four molecular units A,B, A’ and B’, where B and B’ denote two sensing elements (for example two primary antibodies or other suitable sensing proteins) which come close to each other in the presence of the target biomarker. A and A’ denote two proteins which when close together flag the presence of the biomarker. Examples of A and A’ are the two moieties of split protein (such as an enzyme, a bio-luminescent protein or a toxin) which only function when A and A’ come together, or two fluorescent proteins in the context of optical Förster resonance energy transfer (FRET). The hinge is designed (or biased) to be open in the absence of the target (so that false positive signals are very low) , and to open and close frequently in its presence, which accounts for the speed of the switch to respond to real time changes.
The key features of the technology are as follows:
- Very high sensitivity, specificity and precision allowing much lower biomarker concentrations to be measured, or the use of greatly reduced quantities of sample.
- Dynamic/real-time response due to much faster ON/OFF switching in the presence/absence of target, making it capable of tracking temporal and spatial changes in concentrations of target analytes.
- It is very small -typically four to nine hundred residues, facilitating its expression and use within living cells.
- The sensing molecules can be proteins, antibodies, antibody fragments, epitope fragments and antibody mimetics, and can be deployed within the cell, or in vitro assays. Biomarkers include antigens, DNA and RNA nucleic acid sequences.
In the context of ‘diagnostics and visualization,’ the advantages of the invention are :
- The functionality of the switch technology is built into its design as a fusion protein, therefore diagnostics tests are designed to be cheap (ideally less than a euro/dollar), simple, low cost and fast; and can track in real time;
- Multiple sensors can be combined in the same kit to test for multiple biomarkers/analytes – this is also known as multiplexing;
- Potential to replace traditional immunoassays methods such as the enzyme-linked immunosorbent assay (ELISA), particularly in Point-of-Care diagnostic platforms, particularly where lab based measurement are not easy such as emergency care and in the field applications;
- Microscopy of cell and subcellular environments to monitor the spatio-temporal concentrations of analytes.
In the context of Therapeutics, the advantages of the invention are targeted delivery of therapeutics at a cellular level, with applications in drug delivery and chemotherapy.
Commercialization
We are now preparing for the next part of this story, commercialization of our technology. I have received significant help from our university technology transfer office (UCD Nova) in several ways: legal expertise and submission of patents; business training and awareness of the commercialization process, and very recently support from Enterprise Ireland, a government agency that seeks to convert intellectual property and technologies of academic research into the products and services of tomorrow. As a consequence, we have concluded that while biomedical imaging is of great merit from a research perspective, the corresponding commercial market is comparatively small. Moreover, while therapeutics are of great potential value and benefit on a societal level, the safety, regulatory and development path is lengthy and complex. Development of opportunities in the latter two markets will be rigorously pursued only in the long term. We therefore intend to focus of our efforts on diagnostics, in particular the segment aimed at ‘Point of Care’ (POC) and emergency, highly infectious disease and in the field testing biosensor markets. The rationale for this choice of segment is as follows. Currently, simple, robust and cheap (costing close to a euro) sensors do exist, most of which are based on lateral flow devices. However their accuracy is fundamentally limited for a variety of reasons, so that they are of little use for target biomarkers where high precision is required, although immensely useful when high accuracy is not needed. They also generally cannot be used to detect multiple biomarker types simultaneously. In contrast, sensors built around our molecular switch should be deployable in similar and even simpler devices, yet potentially be far more accurate, target multiple biomarkers simultaneously using very small samples (e.g., a drop of blood), and cost the same or even less. While laboratory equipment, largely based on ELISA or similar assays, are highly accurate they cannot be easily deployed for our target market due to the complexity and cost of the equipment. Examples of diseases where very sensitive measurement of biomarkers are good/useful indicators/predictors include: myocardial infarction (i.e., heart attack), sepsis, tuberculosis, stroke and many forms of cancer. That a POC market exists for our technology is based on an analysis of current scientific and commercial reports, and interviews with experts in academia and industry.
We envisage three stages for the follow up commercialization project.
- The design and optimization of one or more commercially relevant demonstration prototypes through advanced molecular simulation, machine learning and mesoscale/multiscale modelling (essentially E-CAM work-packages 1 and 4 respectively) on massively parallel computer platforms , and their subsequent building and and characterization in cell-lines using genetic recombination methods and FRET microscopy by experimental colleagues at University College Dublin.
- The building of nano-fluidic devices to perform homogeneous liquids assays using our protein bases sensors in collaboration with colleagues at the Irish Biomedical Diagnostics Institute who have extensive experience building “labs on a chip” and host a Fraunhofer project centre.
- Design and licensing in collaboration with industry partners of versions of our technology specific to their target markets through a start-up company that we will set up.
And you might ask what happened to the PhD student with whom I this started this story? He did receive his PhD, and now is the CEO of a startup called Think Bio Solutions developing sensors for the sports garment market which allow the wearer to have real time diagnostics and analytics of their physiology as the train. Of course, as a co-inventor of the first patent, he will be entitled to a share of patent royalties, should the sensor prove successful commercially.
Additional Technical Notes
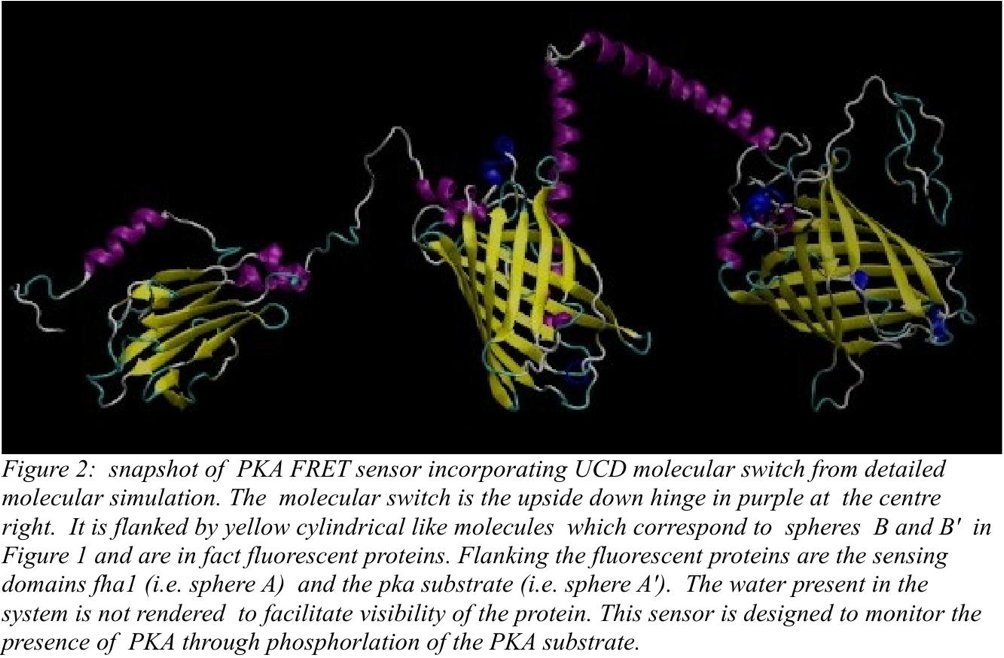
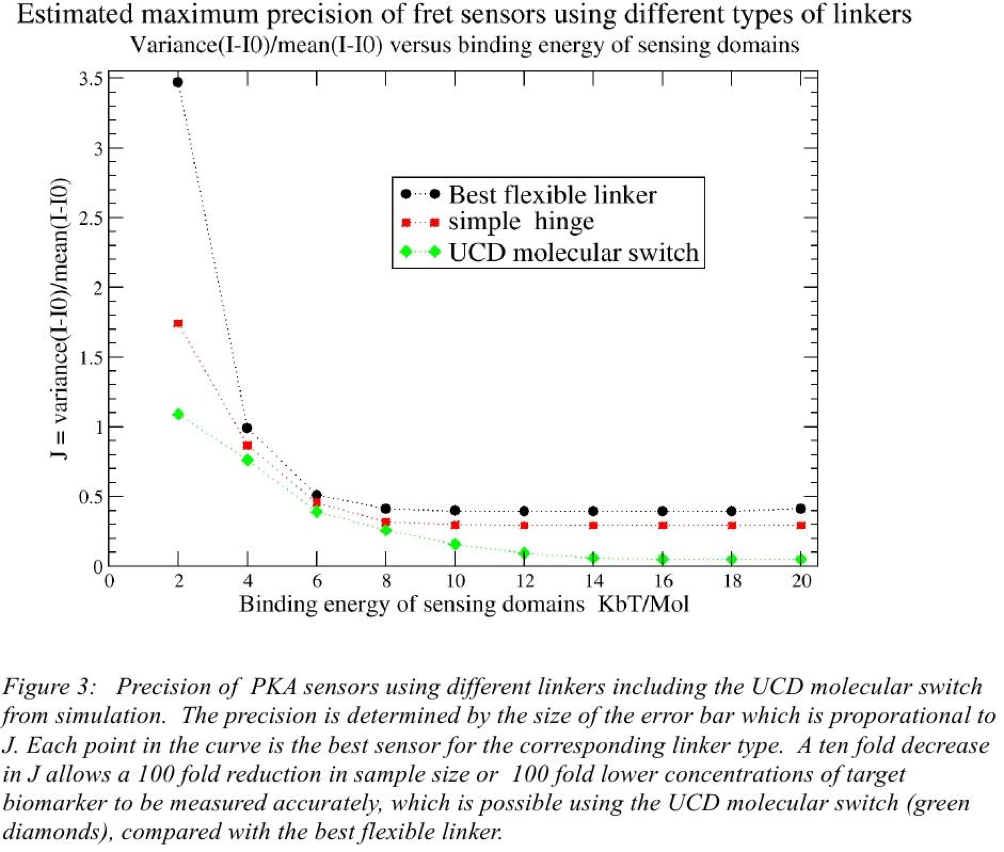
The first system we really focused on was a PKA sensor. PKA otherwise known as protein Kinase A is in fact a family of enzymes whose activity is dependent on cellular levels of cyclic AMP. PKA has several functions in the cell, including regulation of glycogen, sugar, and lipid metabolism. It was discovered by chemists H. Fischer and Edwin G. Krebs in 1968, who won the Nobel Prize in Physiology or Medicine in 1992 for their work on phosphorylation and dephosphorylation and how it relates to protein kinase A activity. A representative example of the application of the molecular switch, monitoring the presence of PKA (protein kinase A) through FRET microscopy, is given in Figure 2. The precision of representative examples of PKA sensors is estimated in Figure 3 through simulation.
Patent Applications
Inventor: D. Mac Kernan PCT/IB2017/055432 (2017) Applicant UCD
Inventors D. Mac Kernan and S. Sanyal PCT/ IB2016/051249 (2016) Applicant UCD
For those interested in knowing more, including possible commercial applications, please contact the researcher directly by e-mail: donal.mackernan@ucd.ie.
