Pascal Carrivain
I did a Ph.D with Jean-Marc Victor (Theoretical physics of condensed matter UMR CNRS 7600)
I did a post-doc with Cédric Vaillant and Ralf Everaers (Laboratoire de Physique, ENS de Lyon)
I am currently an E-CAM post-doc with Ralf Everaers (Centre Blaise Pascal, ENS de Lyon)
Host beneficiary: ENS Lyon, France
Description
To study the long term memory of the initial conformation of a highly entangle polymer we need to preserve the topology. It means that two bonds of the polymer cannot cross. It is of great importance for the study of post-mitotic chromosome unfolding, as-well-as the study of material rheological properties. To resolve the excluded volume constraints (between two polymer bonds) you could use a soft or hard potential between the two points associated to the minimal distance. Here, we propose to change the relative velocity between overlapped bonds to resolve the excluded volume constraint in one time-step of molecular dynamics. I implemented one module to compute correct minimal distance for LAMMPS (molecular dynamics simulator) and one module to resolve excluded volume for LAMMPS. It takes advantage of the underlying parallelism.
The velocity-based method is an algorithm developed by the physics engine field to produce realistic motion in video games ... I adapted the method to the polymer simulation field.
In a second time, I would like to create a python bindings for Open-Dynamics-Engine (open source physics engine). Physics engine can resolve excluded volume constraints for arbitrary articulated system with complex shapes. The python bindings for Open-Dynamics-Engine would be a new engine that can be interfaced with Open-Path-Sampling openpathsampling.org. Of note, I would like to work on modules to speed-up Open-Dynamics-Engine solver computation time.
Development Plan
List of Tasks
- Module 1 : minimal distance between two segments using the Karush-Kuhn-Tucker conditions
- Module 2 : resolve the excluded volume constraints with velocity-based method
- Module 3 : openmm_plectoneme
- Module 4 : openmm_copolymer
- Module 5 : polymer_data_analysis
- Module 6 : Verlet_list_for_ODE
- Module 7 : solve_collisions_first_ODE
- Module 8 : 2spaces_on_gpu
- Module 9 : Python bindings for "Open-Dynamics-Engine" (Work-In-Progress)
List of Modules
Module 1: minDist2segments_KKT
Status: Done
Description: The minDist2segments_KKT module returns the minimal distance between two line segments. It uses the Karush-Kuhn-Tucker conditions (KKT) for the minimization under constraints. To resolve excluded volume constraints one could use a soft or hard potential between the two points associated to the minimal distance. I implemented correct computation of minimal distance between two bonds for LAMMPS. A standalone version exists. The module is used by velocities_resolve_EVC module.
Module 2: velocities_resolve_EVC
Status: Done
Description: Polymer physics uses bead-spring model to ensure connectivity between consecutive monomers and Weeks-Chandler-Andersen (WCA) repulsive potential to avoid bond crossing. The present module proposes to implement a method that comes from physics engine. It uses velocity-based method to resolve the excluded volume constraints in one time-step of molecular dynamics. We can see it as a hard-potential that pushes away two bonds in one time-step while WCA can be seen as a soft potential that needs more time-step to remove/prevent bond overlapping. I adapted the method from 3D rigid body (position and orientation are given) to the bead-spring model where only the position of the beads are known.
The module is used to study genome-wide unfolding while preserving the topology of fruit fly mitotic chromosome. I implemented a standalone version as-well-as a LAMMPS version.
I implemented velocities_resolve_EVC module for LAMMPS.
Description:
Module 3: openmm_plectoneme
Status: Done
Description: The openmm_plectoneme module introduces twist to a ring polymer and sample the accessible conformations under torsionnal constraints.
This module takes advantage of the OpenMM software and GPU acceleration.
It builds a Kremer-Grest polymer model with virtual sites to attach a frame to each bond.
The frame is used to compute twisting forces.
We also implemented a replica exchange in the mechanical space parameters using HPC Dask jobqueue software.
The module is used in a scientific collaboration to study super-braided bacteria conformations.
Module 4: openmm_copolymer
Status: Done
Description: The openmm_copolymer module assists the creation of block-copolymer model and sample accessible conformations.
This module takes advantage of the OpenMM software and GPU acceleration.
It builds a Kremer-Grest polymer model with uni-dimensionnal epigenetic information and construct the epigenetic interactions based on the model you would like to study.
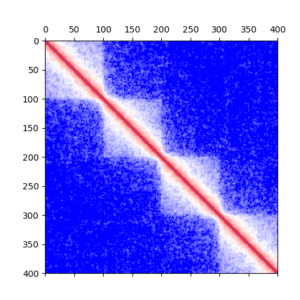
Module 5: polymer_data_analysis
Status: Done
Description: The polymer_data_analysis module provides functions to compute quantities like gyration radius, internal distances, contact maps, contact probabilities, writhe ...
This module takes advantage of the Numba paralleling implementation and just-in-time compiler.
It also uses HPC Dask software to run data analysis on job queuing systems.
It is used to analyze data produces by openmm_plectoneme and openmm_copolymer modules.
Module 6: Verlet_list_for_ODE
Status: Done
Description: Open-Dynamics-Engine is an open source physics engine software.
The software solves mechanical articulated system and collisions between arbitrary complex shapes to produce realistic motion for video games.
The Verlet_list_for_ODE module introduces Verlet-list to speed-up collision detection.
The module provides two examples: one of them is a rigid-body model for bacteria and the other is a rigid-body model of chromosome at the scale of DNA and histones assembly.
Module 7: solve_collisions_first_ODE
Status: Done
Description: Open-Dynamics-Engine software resolves excluded volume constraints for any complex shape and arbitrary articulated system.
The solve_collisions_first_ODE module splits the solver.
First, the module asks for Open-Dynamics-Engine software to resolve the excluded volume constraints and apply it to the articulated system.
Then, the articulated system is solved and the system steps to the next state.
The two steps solver aim is to speed-up the resolution of both excluded volume and articulated constraints.
Module 8: 2spaces_on_gpu
Status: Done
Description: The 2spaces_on_gpu module implements the 2-spaces algorithm on GPU (see Massively Parallel Architectures and Polymer Simulation and Cellular automata for polymer simulation with application to polymer melts and polymer collapse including implications for protein folding for more details about the algorithm).
This algorithm is designed to move one-half of the polymer in one Monte-Carlo iteration.
It also preserves the excluded volume constraints. I wrote a OpenCL implementation to be used on both CPU and GPU. In particular, the code runs at least ten times faster on GPU hardware.