In the last few years, modelling of rare events has made tremendous progress and several computational methods have been put forward to study these events. Despite this effort, new approaches have not yet been included, with adequate efficiency and scalability, in common simulation packages. One objective of the Classical Dynamics Work Package of the project E-CAM is to close this gap. The present text is an easy-to-read article on the use of path sampling methods to study rare events, and the role of the OpenPathSampling package to performing these simulations. Practical applications of rare events sampling and scalabilities opportunities in OpenPathSampling are also discussed.
Rare events
In many simulations, we come across the challenge of bridging timescales. The desire for high resolution in space (and therefore time) is inherently in conflict with the desire to study long-time dynamics. To study molecular dynamics with atomistic detail, we must use timesteps on the order of a femtosecond. However, many problems in biological chemistry, materials science, and other fields involve events that only spontaneously occur after a millisecond or longer (for example, biomolecular conformational changes, or nucleation processes). That means that we would need around 1012 time steps to see a single millisecond-scale event. But when these events do occur, they do so quickly. The event of interest is not slow but rather rare, requiring long waiting times before it is observed.
Two fundamental problems of statistical mechanics are intimately tied to the time scale problem of classical molecular dynamics simulation:
a) The calculation of the populations of metastable states of an equilibrium system. Such populations can be expressed in terms of free energies and hence this problem boils down to the efficient calculation of free energies;
b) The sampling of transition pathways between long-lived (meta)stable states and the calculation of reaction rate constants.
Although the first problem is a static one and does not necessarily require following the dynamics of the system, free energies are often computed using molecular dynamics. Since the understanding of rare but important events also requires the calculation of free energy barriers, which are related to rare configurations, such simulations are affected by the rare event problem. In principle, this problem can be solved by running MD simulations for a very long time. In the best of cases such simulations will be expensive, but often they are simply unfeasible on current or even foreseeable computers. Similarly, rare transitions between long-lived states can be found by running an MD simulation until the transition of interest occurs. However, within the accessible computing time the event may never happen. In the past decades, several powerful algorithms have been developed to overcome the time scale problem both for free energy computation and for rare event sampling. Typically, these methods apply an appropriately constructed bias or constraint, which artificially increases the likelihood of the rare event in a way such that it is possible to correct for the bias and restore the true probability of the event. In contrast to straightforward molecular dynamics, for which a number of excellent software packages are available (e.g., Lammps, Charmm, Gromacs, NAMD, etc.), methods for rare event sampling have not yet been implemented, with the required efficiency and scalability, into widespread simulation packages. One objective of Work Package 1 (WP1) of the project ECAM is to close this gap and develop well tested and robust software modules for free energy computation and rare event sampling.
We redirect the reader to Deliverable D1.1 of WP1, Identification/selection of E-CAM MD codes for development, for an overview of the most widely used algorithms for free energy calculations and the sampling of trajectories involving rare events, and a discussion of the capabilities of currently available software packages that incorporate (some of) these algorithms. In this article, we will focus on the work that E-CAM is doing on point b) above, and the development of trajectory sampling (also called “path sampling”) software modules for studying the thermodynamics and kinetics of rare events.
Path Sampling
While modern supercomputers are beginning to make it possible to obtain trajectories long enough to observe some of the above mentioned rare processes (such as millisecond dynamics of a protein [1]), even then, we may only find one example of a given transition. To fully characterize a transition (with proper statistics), we need many examples. This is where path sampling comes in.
Path sampling approaches obtain many trajectories using a Markov chain Monte Carlo approach: an existing trajectory is perturbed (usually using a variant of the “shooting” move), and the resulting trial trajectory is accepted or rejected according to conditions that preserve the distribution of the path ensemble. As such, path sampling in Monte Carlo is the space of paths (trajectories). Conceptually, this enhances the sampling of transitions by focusing on the transition region instead of the stable states. In direct MD, trajectories spend much more time in stable states than in the transition region (exponential population differences for linear free energy differences); path sampling skips over that time in the stable states (Figure 1).
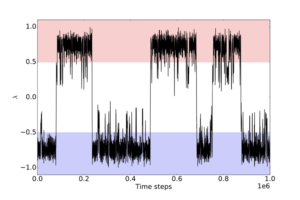
Figure 1: Typical example of a rare event. The x-axis is simulation time, the y-axis is some order parameter that distinguishes two states, coloured in blue and red. Most of the time is spent in one stable state or the other, and transitions between them are rare. Path sampling focuses only on the sharp, nearly vertical jumps where the trajectory makes a transition between the two states. It therefore avoids all the time in between, thus gaining enormous savings in computing effort.
The main path sampling approaches used in the software developed in E-CAM to study rare events are transition path sampling (TPS) [2] and transition interface sampling (TIS) [3]. In practice, TPS is mainly used to characterize the mechanism of a transition, while TIS (which is more expensive than TPS) is used to calculate rates and free energy landscapes. Overviews of these methods, as well as other rare events methods, can be found in recent review articles such as Refs [4] and [5]. Figure 2 is an example of how the transition path sampling methodology allows assessing the long-time dynamics of protein conformational changes.
In addition, several other resources are available on the web to teach path sampling, including:
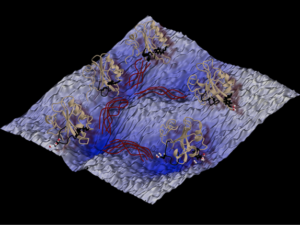
Figure 2: The reaction network of conformation changes of the photo-active yellow protein studied with transition path sampling. In the background is the rough energy landscape with several (free energy) minima, and for each minimum a representative snapshot of the protein conformation is shown in cartoon style. The dark red curves between the minima depict the concept of transition path ensemble, sampled by TPS, and each represents a specific transition state. This picture is a courtesy of Jocelyne Vreede, and is taken from Ref [6].
The OpenPathSampling package
Currently, there are no packages that dominate the field of trajectory-based rare event simulations. Since trajectory-based rare events methods leave the dynamics of the system unchanged, the primary computational cost is in the underlying dynamics engine, and packages that implement these methods usually wrap around some other molecular dynamics package.
As the central difficulty of determining reaction paths between dominant metastable and equilibrium states is still very challenging, and of great importance in biology, chemistry and physics, this has led to an increased focus on the development of codes to fill this gap. OpenPathSampling (OPS) is an effort on that direction, that is built to facilitate path sampling algorithms. Written in Python, OPS can perform TPS and TIS simulations and is independent of the underlying molecular dynamics engine. Currently has support for OpenMM, as well as an internal toy engine, and development is underway to support other MD engines, including LAMMPS and Gromacs.
The team of developers of OPS includes two scientists from E-CAM (Prof. Peter Bolhuis and Dr. David Swenson, from the University of Amsterdam) and the package is still under development for its version 1.0 release. A paper introducing the code is currently in preparation for publication [7], and the website is http://openpathsampling.org/. Overall, OPS is ready for production work and version 1.0 will be released once the paper about the code has been accepted for publication.
The basic tools for several rare event methods, including TPS and TIS, are already implemented in OPS, and it also supports advanced techniques such as variants of replica exchange TIS. It has a generic approach to sampling path ensembles, which makes it very easy to add new methodologies. In addition, OPS supports various underlying molecular dynamics engines. Its external engine module launches the external engine once, and reads the trajectory from the file system during the simulation. It then kills the trajectory when it reaches a stopping point, thus reducing the costs of restarts and overshooting. On the other hand, if the engine provides a direct Python interface (for example, OpenMM), OpenPathSampling uses that instead, and therefore never overshoots. Furthermore, OpenPathSampling also includes extensive unit tests and in-code documentation, as the E-CAM software standards require.
The path sampling software modules developed in E-CAM are based on the OpenPathSampling software package, and several play fundamental roles in this package. Documentation on some of the software development projects within the OPS package can found in E-CAM’s WP1 software repository. A report was also build around these codes in deliverable D1.2 Classical MD E-CAM Modules I, which is accessible from here.
Practical Applications of rare events sampling
Since the problem of bridging timescales, which path sampling addresses, is a generic one, path sampling can be used in many fields. Indeed, there’s nothing in the methodology that even restricts it to molecular simulation. However, it is best known in the field of classical MD simulations, where path sampling methods have shown many successes, including:
- Mechanisms of complex chemical reactions, such as autoionization of water [8]
- Mechanism of hydrophobic assembly [9]
- Evidence that the glass transition is a first-order phase transition [10]
- Mechanism of crystal nucleation [11]
- Mechanism of cavitation [12]
- Identifying new mechanisms in catalytic systems [13]
- Characterization of the conformational dynamics networks in proteins [14]
As computational resources become more powerful, path sampling has the promise to provide insight into rare events in larger systems, and into events with even longer timescales. For example:
-
Drug/protein binding and unbinding (timescales of minutes), which is essential for predicting the efficacy of drugs. The pilot project being performed by Dr. Swenson in collaboration with BiKi Technologies, an E-CAM industrial partner, focuses on this problem in the case of a particular drug and its target protein.
-
Association processes of proteins (large systems), which is at the core of communication in biochemical pathways.
-
Self-assembly processes for complex systems (many intermediates), which can be important for the design of new materials.
Further, applying the known successes of path sampling methods to larger systems can also be quite valuable. Path sampling can shed light on the networks of conformational dynamics for large proteins and protein complexes, and on the mechanisms and rates of complex reactions and phase transitions. The range of possibilities is so broad that it is impossible to enumerate – both academics and industry will benefit greatly from having software for these methods.
Scalability opportunities
Software for trajectory-based rare events simulations, such as OpenPathSampling, usually wrap around other MD engines, because these simulations don’t change the underlying dynamics. This gives them the potential for a wider range of applications. Furthermore, since the primary cost of the calculation is in performing the dynamics, this also means that most of the responsibility for performance falls on the underlying engine. OPS already has support for OpenMM, a GPU-accelerated MD code, and support is in development for the widely-used and highly-scalable packages LAMMPS and Gromacs. Each of these dynamics engines is designed for high performance computing, and in this way, OPS inherits the performance from the underlying dynamics code. This approach means that as new generations of hardware lead to new molecular dynamics codes or changes in existing codes, OpenPathSampling can continue to use the cutting edge in hardware with minimal modification.
Bibliography
[1] Kresten Lindorff-Larsen, Paul Maragakis, Stefano Piana, and David E. Shaw. Picosecond to millisecond structural dynamics in human ubiquitin. J. Phys. Chem. B, 120(33):8313, 2016.
[2] Christoph Dellago, Peter G Bolhuis, Félix S Csajka, and David Chandler. Transition path sampling and the calculation of rate constants. J. Chem. Phys., 108(5):1964, 1998.
[3] Titus S van Erp, Daniele Moroni, and Peter G Bolhuis. A novel path sampling method for the calculation of rate constants. J. Chem. Phys., 118(17):7762, 2003.
[4] Peter G Bolhuis and Christoph Dellago. Trajectory-Based Rare Event Simulations. In Reviews in Computational Chemistry, Volume 27, pages 111–210. John Wiley & Sons, Inc., Hoboken, NJ, USA, 2010.
[5] Christoph Dellago and Peter G Bolhuis. Transition Path Sampling and Other Advanced Simulation Techniques for Rare Events. In Advanced Computer Simulation Approaches for Soft Matter Sciences III, pages 167–233. Springer, Berlin, Heidelberg, Berlin, Heidelberg, 2008.
[6] Jarek Juraszek, Jocelyne Vreede and Peter G Bolhuis. Transition path sampling of protein conformational changes. Chemical Physics, 396 (2012) 30 – 44
[7] Jan-Hendrik Prinz, David W.H. Swenson, Peter G. Bolhuis, and John D. Chodera. OpenPathSampling: A Python framework for path sampling simulations. I. Introduction and usage. In prep.
[8] Phillip L Geissler, Christoph Dellago, David Chandler, Jürg Hutter, and Michele Parrinello. Auto ionization in Liquid Water. Science, 291(5511):2121, 2001.
[9] Adam P Willard and David Chandler. The role of solvent fluctuations in hydrophobic assembly. J. Phys. Chem. B, 112(19):6187, 2008.
[10] L O Hedges, R L Jack, J P Garrahan, and D Chandler. Dynamic Order-Disorder in Atomistic Models of Structural Glass Formers. Science, 323(5919):1309–1313, 2009.
[11] Wolfgang Lechner, Christoph Dellago, and Peter G Bolhuis. Role of the Pre structured Surface Cloud in Crystal Nucleation. Phys. Rev. Lett., 106(8):085701, 2011.
[12] Georg Menzl, Miguel A Gonzalez, Philipp Geiger, Frédéric Caupin, José L F Abascal, Chantal Valeriani, and Christoph Dellago. Molecular mechanism for cavitation in water under tension. PNAS, 113(48):13582, 2016.
[13] Cynthia S Lo, Ravi Radhakrishnan, and Bernhardt L Trout. Application of transition path sampling methods in catalysis: A new mechanism for CC bond formation in the methanol coupling reaction in chabazite. Catalysis Today, 105(1):93, 2005.
[14] Jocelyne Vreede, Jarek Juraszek, and Peter G Bolhuis. Predicting the reaction coordinates of millisecond light induced conformational changes in photoactive yellow protein. Proc. Natl. Acad. Sci. U.S.A., 107(6):2397, 2010.
